
1. 데이터 다운로드
- 링크 : https://github.com/mibwurrepo/Microbial-bioinformatics-introductory-course-Material-2018
- 다운 방법 : 링크 접속 -> code -> Download ZIP
- 분석 환경 세팅
- Download ZIP클릭
- [the Microbial-bioinformatics-introductory-course-Material-2018-master.zip] 폴더 압축 풀기
- [input_data] 압축 풀기
- [humanmicrobiome.zip] 압축 풀기 -> 폴더 안에 있는 "humanmicrobiome.biom"를 [input_data] 폴더로 옮기기
- "microbiometutorials.Rproj" 파일 클릭 -> 현재 위치를 기본 위치로 설정해 준다
2. 데이터 살펴보기
ps <- readRDS("./phyobjects/ps.ng.tax.rds")
# phyloseq-class experiment-level object
# otu_table() OTU Table: [ 4710 taxa and 474 samples ]
# sample_data() Sample Data: [ 474 samples by 30 sample variables ]
# tax_table() Taxonomy Table: [ 4710 taxa by 6 taxonomic ranks ]
# phy_tree() Phylogenetic Tree: [ 4710 tips and 4709 internal nodes ]474개의 샘플과 그 샘플에 관한 30가지의 데이터와 6개의 랭크를 가진 계통수를 포함하고 있다
phyloseq데이터를 만드는 방법은 원문의 4.4 Making a phyloseq object를 참고 바람
otu_table(ps)[1, ]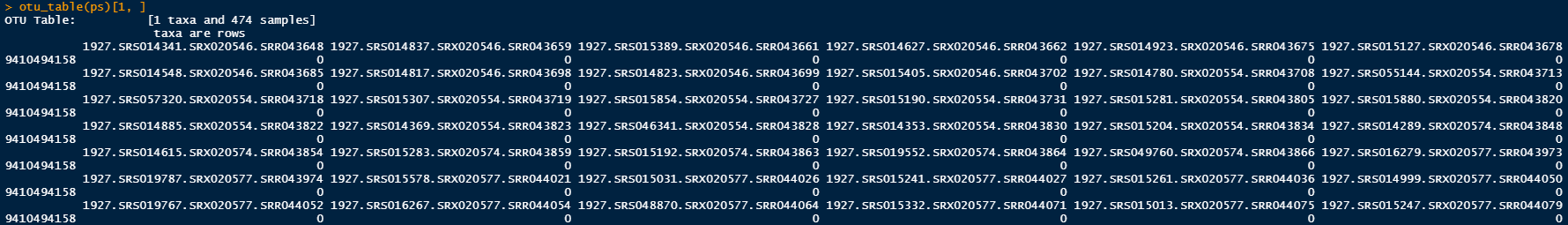
3. 패키지 다운로드 및 phyloseq 파일 import
# import phyloseq
ps <- readRDS("./phyobjects/ps.ng.tax.rds")
# 데이터가 너무 크니가 크기를 줄이자
ps_rs = prune_taxa(taxa_sums(ps) > 100, ps)
ps_prop <- transform_sample_counts(ps_rs, function(x) 100*x/sum(x)) #상대도수로 바꾼다
4. Sample별로 NMDS 분석 plot그리기
이제 원하는 분석을 해준다
otu <- otu_table(ps_prop)
ord <- ordinate(physeq = ps_prop,
method = "NMDS", # Default is "DCA" or "CCA", "RDA", "CAP", "DPCoA", "NMDS", "MDS", "PCoA"
distance = "bray") # Default is "bray" or "unifrac", "wunifrac", "jaccard",
p <- plot_ordination(ps_prop, ord, "samples", color="sample_type")
p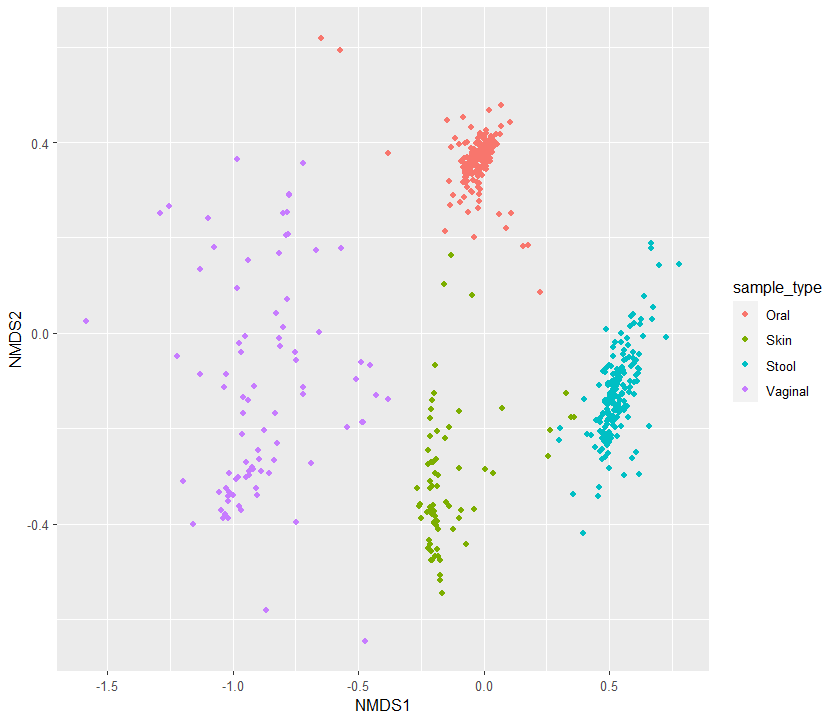
5. ggplot으로 그리기
samples <- data.frame(scores(ord$points))
samples$SampleType<- get_variable(ps_prop, "sample_type") # sampledata에 "sample_type"이라는 열을 가져와서 열 추가
samples
# MDS1 MDS2 SkinType
#1927.SRS014341.SRX020546.SRR043648 -0.81474818 0.16662040 Vaginal
#1927.SRS014837.SRX020546.SRR043659 -1.05460391 -0.33291868 Vaginal
#1927.SRS015389.SRX020546.SRR043661 0.57928105 -0.05124357 Stool
#1927.SRS014627.SRX020546.SRR043662 -0.92117159 -0.28196423 Vaginal
#1927.SRS014923.SRX020546.SRR043675 0.47449253 -0.16889302 Stool
#1927.SRS015127.SRX020546.SRR043678 -0.05396112 0.34111887 Orallibrary(ggplot2)
p <- ggplot(samples) +
geom_point(aes(x = MDS1, y = MDS2, color=SampleType))
p같은 그림이 나온다
stat_ellipse()를 사용해 원으로 묶어 주자
ggplot(samples)+ #sets up the plot. brackets around the entire thing to make it draw automatically
geom_point(aes(x = MDS1, y = MDS2, color=SampleType)) + #puts the site points in from the ordination, shape determined by site, size refers to size of point
stat_ellipse(level = 0.9, aes(x = MDS1, y = MDS2, fill=SampleType),geom = "polygon", alpha = 0.3)+
theme_bw()+
theme(legend.position = "none")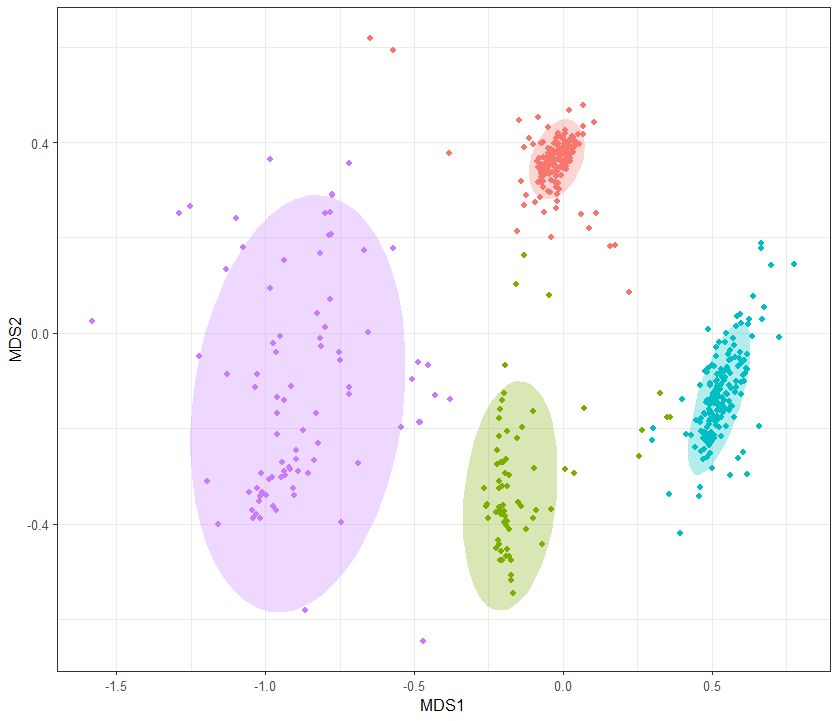
6. phylum 별로 NMDS 분석 plot그리기
spps <- as.data.frame(scores(ord, choices=c(1,2),display=c("species")))
spps_species <- as.data.frame(tax_table(ps_prop))
head(spps_species)
# Domain Phylum Class Order Family Genus
#9410494158 Bacteria Bacteroidetes Bacteroidia Bacteroidales Prevotellaceae Prevotella
#9410494576 Bacteria Bacteroidetes Bacteroidia Bacteroidales Prevotellaceae Prevotella
#9410491420 Bacteria Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides
#9410491569 Bacteria Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides
#9410491595 Bacteria Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides
#9410491783 Bacteria Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides
spps$Phylum <- factor(spps_species$Phylum) # making a column with species names
head(spps)
# NMDS1 NMDS2 Phylum
#9410491526 0.6314447 -0.08904021 Bacteroidetes
#9410491516 0.6231495 -0.10382150 Bacteroidetes
#9410492612 0.4698253 -0.37411464 Bacteroidetes
#9410491521 0.6435790 -0.04185019 Bacteroidetes
#9410491824 0.5839583 -0.02245883 Bacteroidetes
#9410491817 0.5585791 -0.16516024 Bacteroidetesp <- ggplot()+
geom_point(data=spps, aes(y=NMDS2, x=NMDS1, color=Phylum))
p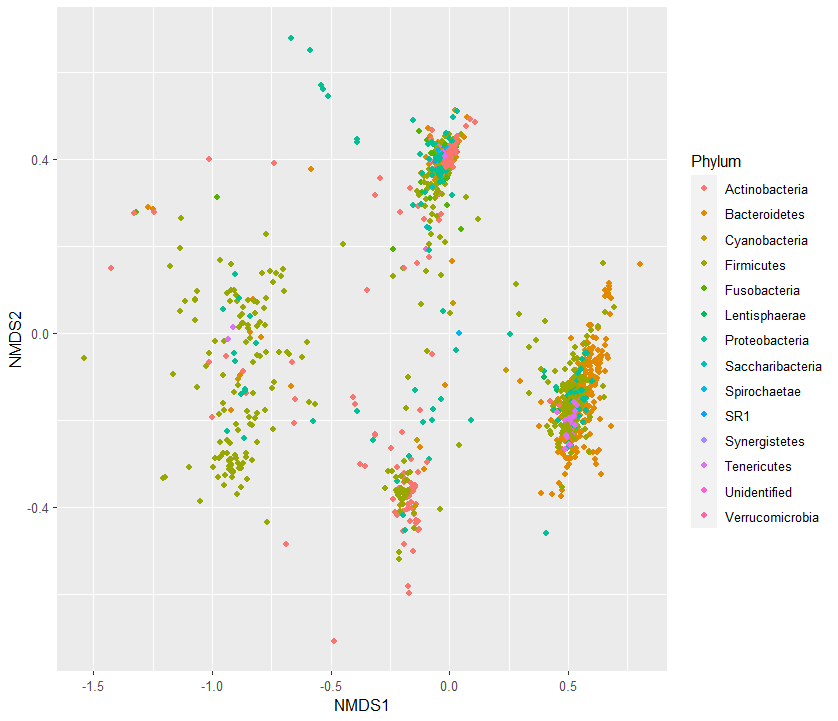
6. phylum + Sample type 별로 NMDS 분석 plot그리기
p <- ggplot(samples)+ #sets up the plot. brackets around the entire thing to make it draw automatically
geom_point(aes(x = MDS1, y = MDS2, shape=SampleType)) + #puts the site points in from the ordination, shape determined by site, size refers to size of point
geom_point(data=spps, aes(y=NMDS2, x=NMDS1, color=Phylum))
p
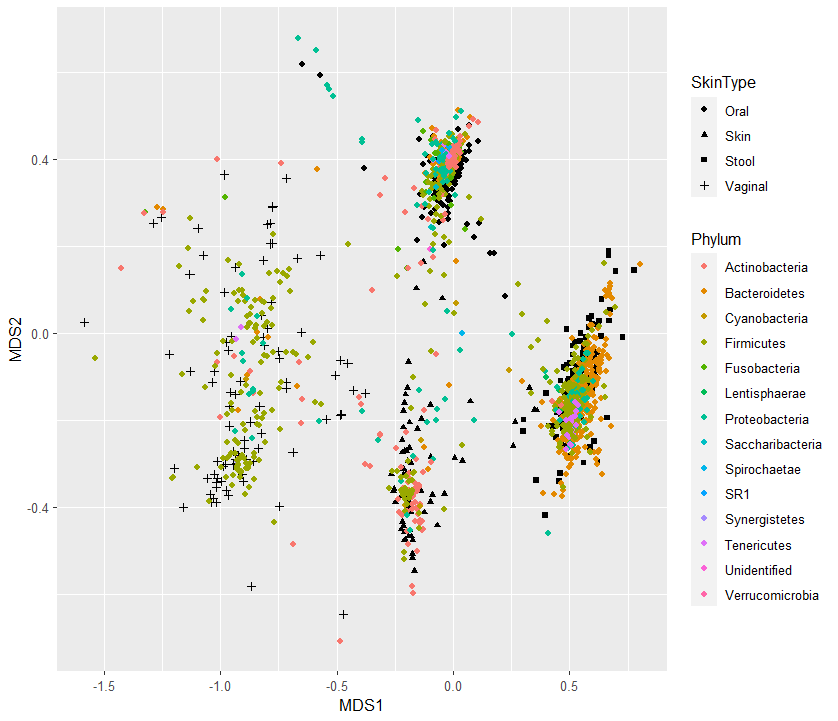
참고
https://github.com/joey711/phyloseq/issues/105
반응형
